New multi-agent system built on Gemini 2.0 accelerates discovery, but human oversight remains critical.
In a bid to accelerate scientific discovery, Google researchers have unveiled an AI "co-scientist" that autonomously generates biomedical hypotheses, debates their merits, and iteratively refines them—a process culminating in lab-validated findings, including potential drug candidates for acute myeloid leukemia (AML) and novel targets for liver fibrosis. The system, detailed in a preprint paper co-authored by teams from Stanford, Imperial College London, and Houston Methodist, represents one of the most advanced attempts yet to integrate large language models (LLMs) into the scientific method itself.
The Architecture: A Self-Improving Hypothesis Engine
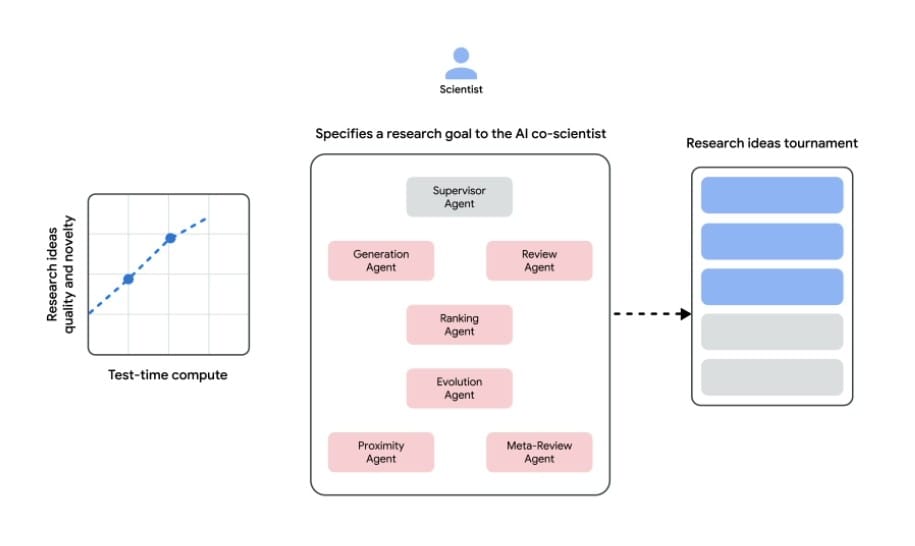
At its core, the co-scientist is a multi-agent framework built on Gemini 2.0, Google’s flagship multimodal model. Unlike earlier AI tools that simply summarize literature or predict protein structures, this system employs specialized "agents" to mimic stages of human scientific reasoning:
- Generation Agent: Scours 33 million PubMed articles and clinical databases to propose initial hypotheses.
- Reflection Agent: Acts as a peer reviewer, fact-checking claims against existing studies and flagging contradictions.
- Tournament System: Uses Elo-style rankings to pit hypotheses against each other in simulated debates, prioritizing novelty and feasibility.
- Evolution Agent: Iteratively refines top candidates by combining ideas or introducing "out-of-the-box" twists.
The system scales computational resources dynamically, allocating more power to promising avenues—a process researchers call "test-time compute amplification." In benchmarks across 15 expert-curated biomedical challenges, this approach outperformed standalone LLMs like GPT-4o and DeepSeek-R1, achieving a 78.4% accuracy rate on the GPQA "diamond set" of complex scientific questions.
From Silicon to Petri Dish: Validating the AI’s Predictions
The team tested the co-scientist on three high-stakes biomedical problems:
-
Drug Repurposing for AML
- Tasked with identifying FDA-approved drugs that could inhibit AML cell lines, the AI proposed Binimetinib (a MEK1/2 inhibitor) and KIRA6 (an IRE1α inhibitor).
- Lab tests showed both compounds suppressed tumor growth at clinically relevant concentrations (IC50 as low as 7 nM for Binimetinib in MOLM-13 cells).
-
Liver Fibrosis Target Discovery
- The AI pinpointed three epigenetic modifiers linked to myofibroblast formation. Two of the corresponding inhibitors reduced fibrogenesis by 40-60% in human hepatic organoids.
-
Antimicrobial Resistance (AMR) Mechanisms
- In a blind test, the system independently rediscovered how capsid-forming phage-inducible chromosomal islands (cf-PICIs) spread antibiotic resistance genes—matching unpublished findings from a decade-long study.
"These aren’t just theoretical proposals," said co-lead author Wei-Hung Weng. "We’re talking about hypotheses that held up under in vitro validation, some within 48 hours of generation."
The Fine Print: Limitations and Ethical Guardrails
Despite its promise, the co-scientist has critical limitations:
- Literature Blind Spots: Relies on open-access papers, missing paywalled studies or negative results rarely published.
- Hallucination Risks: While safeguards filter 92% of flawed proposals, the system occasionally "invents" supporting data for hypotheses.
- Narrow Focus: Validated only in biomedicine; performance in physics or materials science remains untested.
To mitigate misuse, Google implemented a "Frontier Safety Framework" that blocks research goals deemed dual-use or unethical. Early red-teaming exercises with 1,200 adversarial prompts successfully triggered zero unsafe outputs, though researchers caution that determined bad actors might still exploit gaps.
The Bigger Picture: Augmentation, Not Automation
The co-scientist aligns with a broader industry shift toward "AI lab partners." Rivals like OpenAI’s Deep Research and Meta’s Galactica have explored similar territory, but Google’s emphasis on wet-lab validation sets this work apart.
Yet for all its computational firepower, the system’s creators stress it’s a tool—not a replacement. The best results may come from human-AI collaboration. Researchers guide the AI’s exploration, interpreted results, and designed follow-ups. That synergy is likely irreplaceable.
What’s Next?
Google is allowing early access to co-scientist through a Trusted Tester Program. But as LLMs increasingly blur the line between assistant and colleague, the study underscores a pressing question: How do we measure—and ethically govern—AI’s role in the scientific canon?
For now, the answer lies in the Petri dishes, where hypotheses born from silicon are stress-tested in the messy reality of biology.
Read the full preprint: Towards an AI Co-Scientist | Related: AlphaFold’s Protein Revolution